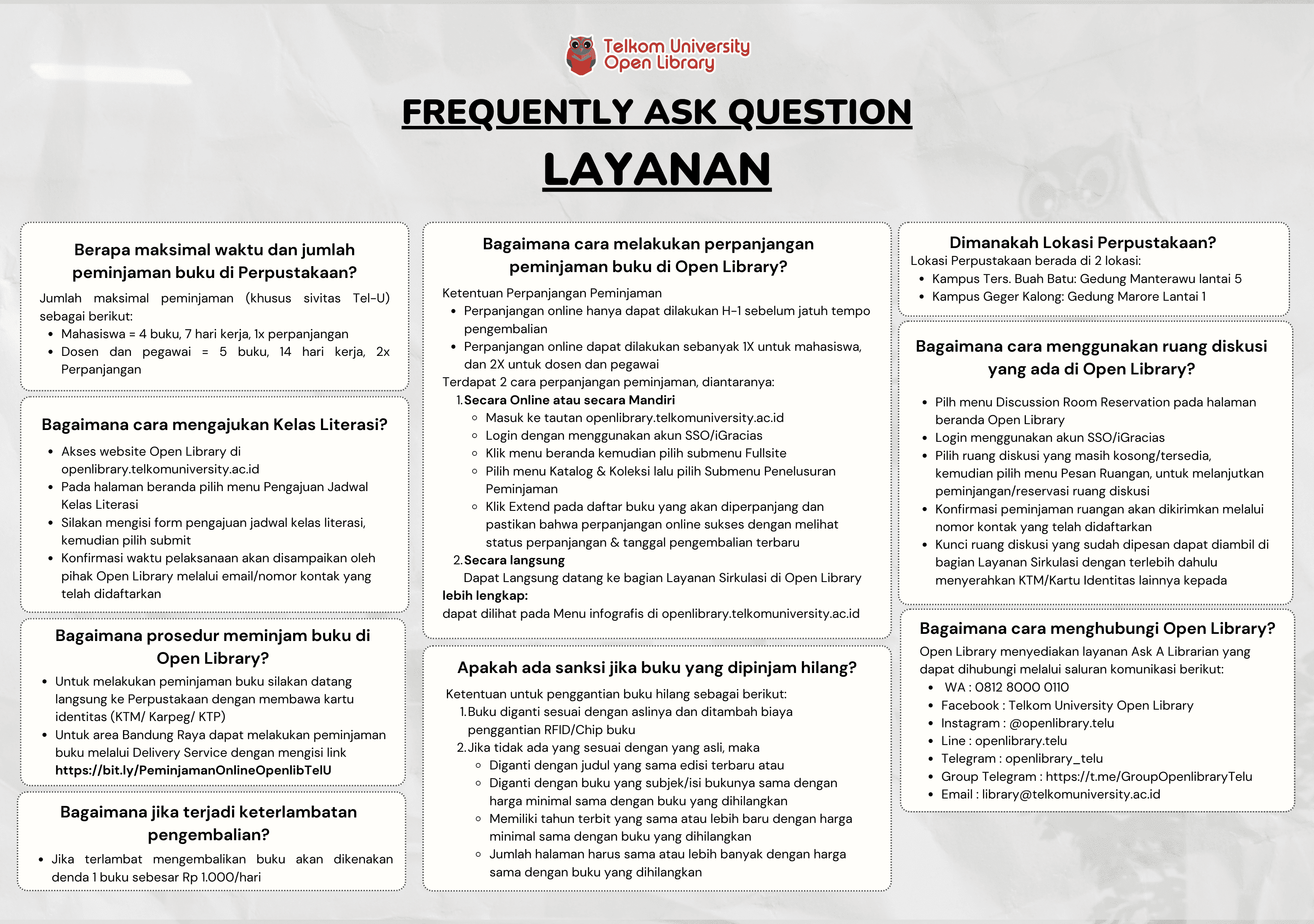
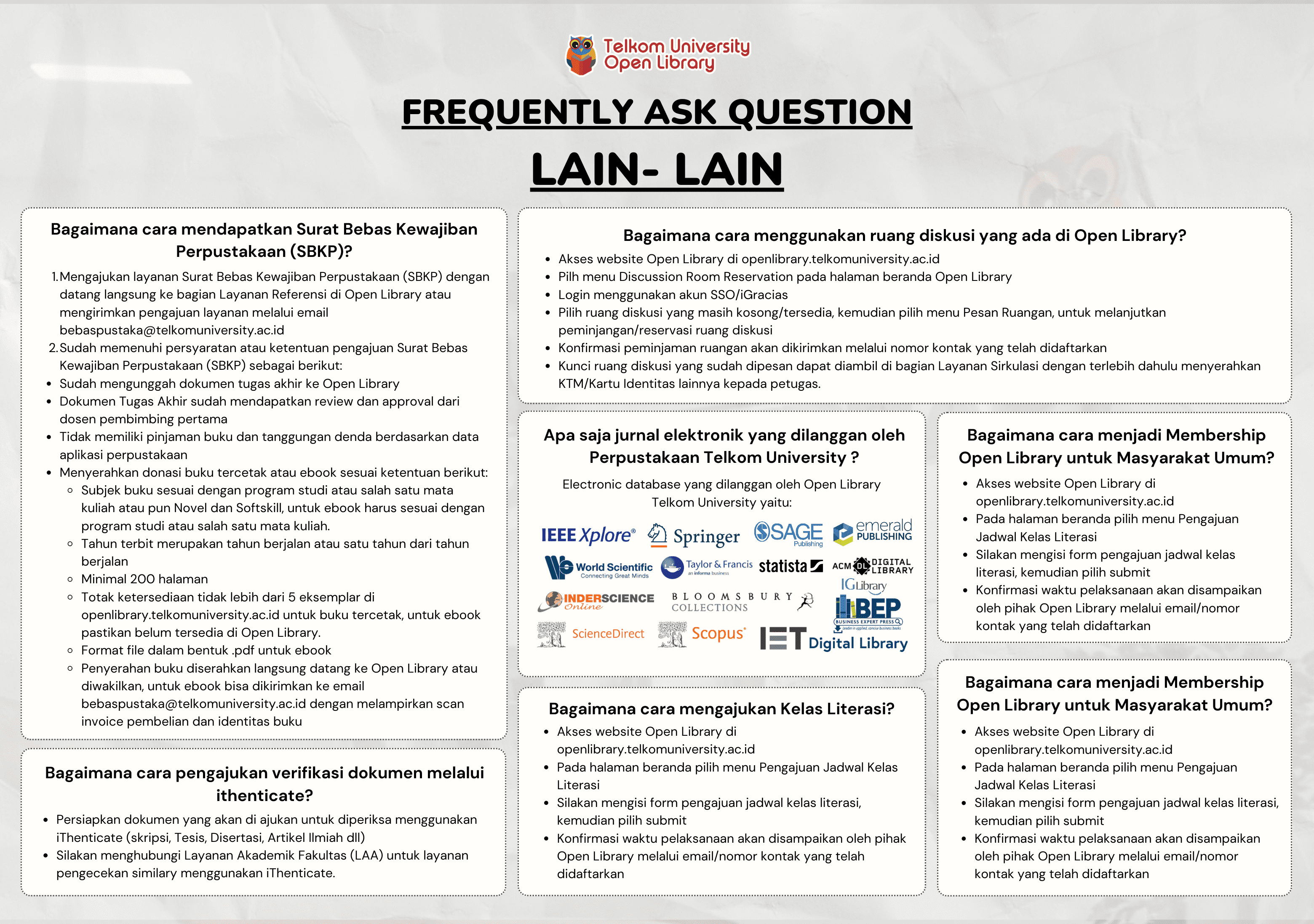
Prediction of Cathepsin K Inhibitors Bioactivity for Bone Diseases Treatment by using Long Short-Term Memory Optimized by Simulated Annealing - Dalam bentuk buku karya ilmiah
ALFIANSYAH HAFIDZ ARBI PUTRA

Informasi Umum
Kode
25.04.7129
Klasifikasi
610.28 - Biomedical Engineering
Jenis
Karya Ilmiah - Skripsi (S1) - Reference
Subjek
Bioinformatics
Dilihat
22 kali
Informasi Lainnya
Abstraksi
<p>Osteoporosis, a bone disease affecting over 200 million people worldwide, presents a significant therapeutic challenge, with Cathepsin K (CatK) being a primary target for inhibitor development due to its role in bone resorption. While conventional drug discovery methods are often slow and costly, machine learning offers a promising alternative. This study addresses the need for more accurate predictive models by developing a robust framework for assessing CatK inhibitor bioactivity. A Long Short-Term Memory (LSTM) network, chosen for its proficiency in handling complex sequential data typical of molecular structures, was optimized using a Simulated Annealing (SA) metaheuristic. The model was trained on a dataset of 1568 molecules from the ChEMBL database, with bioactivity classified based on <!--[if gte msEquation 12]><m:oMath><b style='mso-bidi-font-weight:normal'><i style='mso-bidi-font-style:normal'><span lang=EN-US style='font-size:10.0pt;font-family:"Cambria Math",serif; mso-fareast-font-family:SimSun;mso-bidi-font-family:"Times New Roman"; mso-ansi-language:EN-US;mso-fareast-language:EN-US;mso-bidi-language:AR-SA'><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>pIC</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>??</m:r></span></i></b></m:oMath><![endif]--> ? values into four categories: Potent (<!--[if gte msEquation 12]><m:oMath><b style='mso-bidi-font-weight:normal'><i style='mso-bidi-font-style:normal'><span lang=EN-US style='font-size:10.0pt;font-family:"Cambria Math",serif; mso-fareast-font-family:SimSun;mso-bidi-font-family:"Times New Roman"; mso-ansi-language:EN-US;mso-fareast-language:EN-US;mso-bidi-language:AR-SA'><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>pIC</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>??</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr> ? </m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>9</m:r></span></i></b></m:oMath><![endif]--> ), Active <!--[if gte msEquation 12]><m:oMath><b style='mso-bidi-font-weight:normal'><i style='mso-bidi-font-style:normal'><span lang=EN-US style='font-size:10.0pt;font-family:"Cambria Math",serif; mso-fareast-font-family:SimSun;mso-bidi-font-family:"Times New Roman"; mso-ansi-language:EN-US;mso-fareast-language:EN-US;mso-bidi-language:AR-SA'><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>(</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>9</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr> > </m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>pIC</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>??</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr> ? </m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>7</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>)</m:r></span></i></b></m:oMath><![endif]--> , Intermediate <!--[if gte msEquation 12]><m:oMath><b style='mso-bidi-font-weight:normal'><i style='mso-bidi-font-style:normal'><span lang=EN-US style='font-size:10.0pt;font-family:"Cambria Math",serif; mso-fareast-font-family:SimSun;mso-bidi-font-family:"Times New Roman"; mso-ansi-language:EN-US;mso-fareast-language:EN-US;mso-bidi-language:AR-SA'><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>(</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>7</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr> > </m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>pIC</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>??</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr> ? </m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>6</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>)</m:r></span></i></b></m:oMath><![endif]--> , and Inactive <!--[if gte msEquation 12]><m:oMath><b style='mso-bidi-font-weight:normal'><i style='mso-bidi-font-style:normal'><span lang=EN-US style='font-size:10.0pt;font-family:"Cambria Math",serif; mso-fareast-font-family:SimSun;mso-bidi-font-family:"Times New Roman"; mso-ansi-language:EN-US;mso-fareast-language:EN-US;mso-bidi-language:AR-SA'><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>(</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>pIC</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>??</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr> < </m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>6</m:r><m:r><m:rPr><m:scr m:val="roman"/><m:sty m:val="bi"/></m:rPr>)</m:r></span></i></b></m:oMath><![endif]--> . The SA-optimized LSTM model significantly outperformed three baseline LSTM models, which achieved a peak average accuracy of 0.77. The optimal SA-tuned configuration (the col_rate95 scheme) attained an average accuracy and F1-score of 0.81. Notably, the model demonstrated exceptional performance in identifying Potent inhibitors, achieving an F1-score of 0.92. However, a key limitation was the difficulty in distinguishing between the Active and Intermediate classes, where misclassifications were more frequent. This research highlights the effectiveness of the SA-LSTM approach in accelerating the discovery of high-bioactivity compounds for osteoporosis treatment. Future work could focus on enhancing model robustness by integrating additional molecular descriptors or exploring alternative deep learning architectures to improve classification accuracy.</p>
Koleksi & Sirkulasi
Tersedia 1 dari total 1 Koleksi
Anda harus log in untuk mengakses flippingbook
Pengarang
| Nama | ALFIANSYAH HAFIDZ ARBI PUTRA |
| Jenis | Perorangan |
| Penyunting | Isman Kurniawan |
| Penerjemah |
Penerbit
| Nama | Universitas Telkom, S1 Informatika |
| Kota | Bandung |
| Tahun | 2025 |
Sirkulasi
| Harga sewa | IDR 0,00 |
| Denda harian | IDR 0,00 |
| Jenis | Non-Sirkulasi |